Markov Lab
Markov Lab
Welcome to the Markov Lab
 The main focus of our research is regulation of mitochondrial gene expression in fungi, specifically, pathogenic yeast species, including Candida albicans and Candida auris.
The main focus of our research is regulation of mitochondrial gene expression in fungi, specifically, pathogenic yeast species, including Candida albicans and Candida auris.
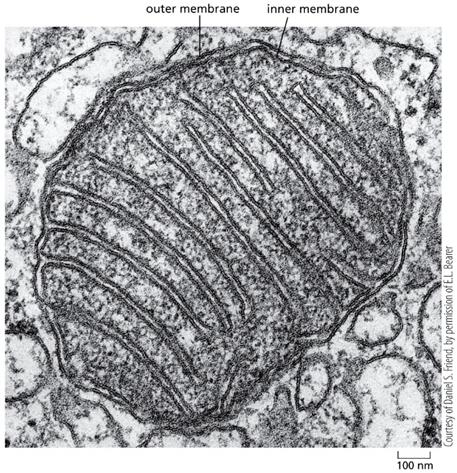
Mitochondria are ubiquitous and amazingly plastic eukaryotic organelles that derive energy from the food by oxidizing organic sources with molecular oxygen through the highly effective process called oxidative phosphorylation and thuosxidasuptivpely cells with vast amounts of ATP, a universal biochemical fuel that drives the cellular metabolism. Mitochondria originate from ancient anaerobic bacteria, and, along with the nucleus, possess their own genome, which expression is driven by enzymes and accessory proteins that are highly homologous to those found in modern bacteria and even viruses. Unlike the nuclear gene expression, wwhhichich is primarily regulated at the initial step – RNA synthesis or transcription, the mitochondrial gene expression
is mostly controlled on post-transcriptional level through the maturation of newly synthesized transcripts, regulation of their stability and availability of protein-encoding RNAs for translation through the interaction with specific auxiliary proteins.
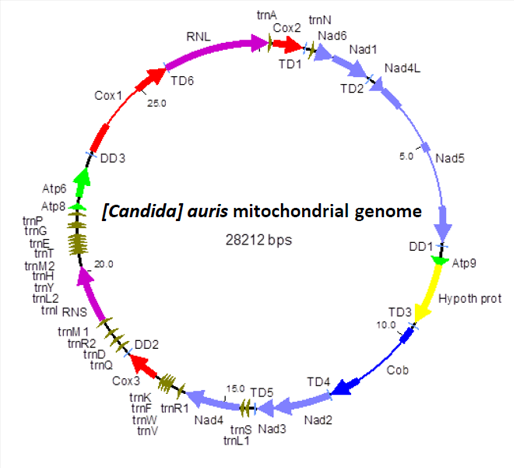
Due to the apparent complexity of aforementioned processes, drastically different organization of fungal mitochondrial genomes in comparison to mitochondrial genomes of animals and plants, and the limited homology between fungal mitochondrial proteins and the mitochondrial proteins of other eukaryotes, the molecular mechanisms that govern mitochondrial gene expression in yeast are still poorly understood. Since the perturbations of mitochondrial metabolism in pathogenic yeast directly influence the outcomes of
anti-fungal drug treatments, the resistance of pathogens to host immune response and their ability to form biofilms, understanding of such mechanisms is crucial for the discovery of potential drug targets that have no similarity to human proteins and thus the development of new anti-fungal therapies.
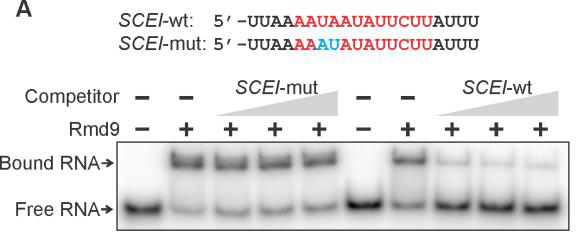
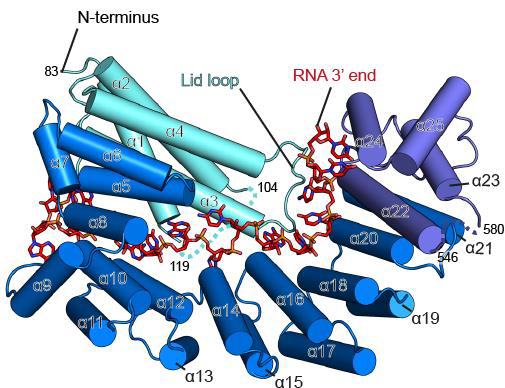
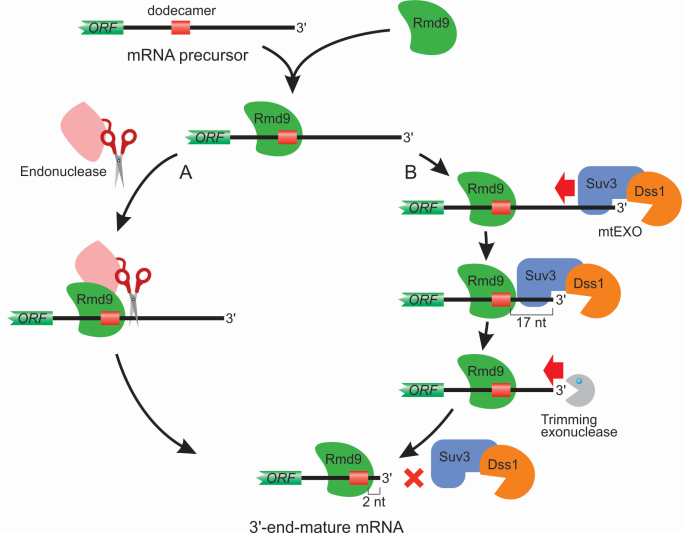
Recently, in close collaboration with Dr. Anikin’s lab (Rowan SOM), we have identified a yeast-specific protein Rmd9 as a key regulator of stability and processing of the 3’ ends of mitochondrial mRNAs (Hillen et al, 2022). It belongs to an ancient family of so-called pentatricopeptide repeat (PPR) proteins that form RNA recognition motifs through the set of
parallel alpha helixes, the tips of which form RNA-interacting interface: PPR proteins are involved in all aspects of yeast mitochondrial gene synthesis, and their loss of function often leads to a complete disruption of respiratory activity in yeast. Despite our understanding of their general architecture, the cellular functions of PPR proteins and the molecular mechanisms of their action remain elusive.
Our current model, which is based on the studies on non-pathogenic yeast Saccharomyces cerevisiae, suggests the involvement of additional factors in the processing of 3’ end of yeast mitochondrial transcripts. In our lab, we use a variety of biochemical, genetic, molecular and cell biology approaches,
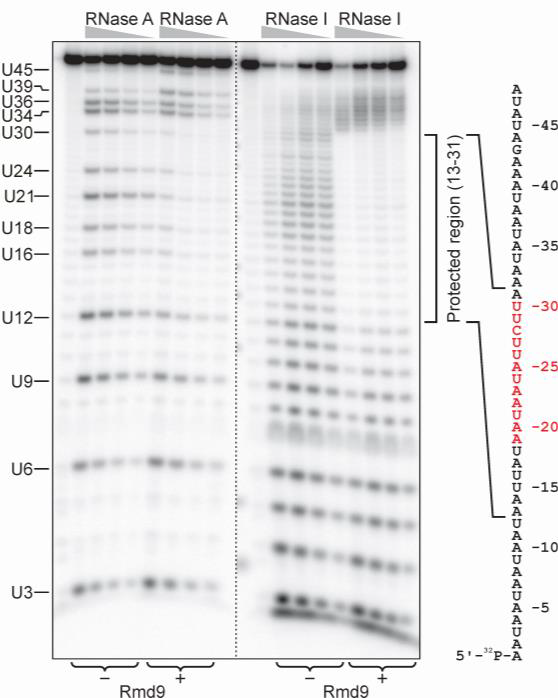
including RNA-protein ‘footprinting’, electrophoresis mobility shift assays (EMSA), fluorescence in vivo, protein and organelle isolation, target-specific pulldowns, high resolution chromatography and mass-spectrometry to further elucidate the details of how this complex system works and to identify its functional components.