Biophysics Projects
Biophysics Projects
Biophysics projects
We study how the voltage sensing phosphatase (VSP) converts an electrical signal into a chemical signal. Our past work looked at how all the different parts of VSP work together. VSP is made up of a voltage sensing domain (VSD), a phosphatase domain and a linker loop that connects the two. Interestingly, the phosphatase domain contains two subdomains, a catalytic domain and a C2 domain. Each region was found to have its own function.
VSDs are made up of four transmembrane helixes, labeled S1-S4. The charge carrying arginines responsible for sensing voltage are found on the S4 (Figure 1). Our results showed that the VSD changes conformation during the depolarizations that turn on the enzyme (1). The conformational changes are similar to those observed from VSDs on ion channels (2). This means that the vast amount of research on ion channel VSDs likely applies to VSP as well.
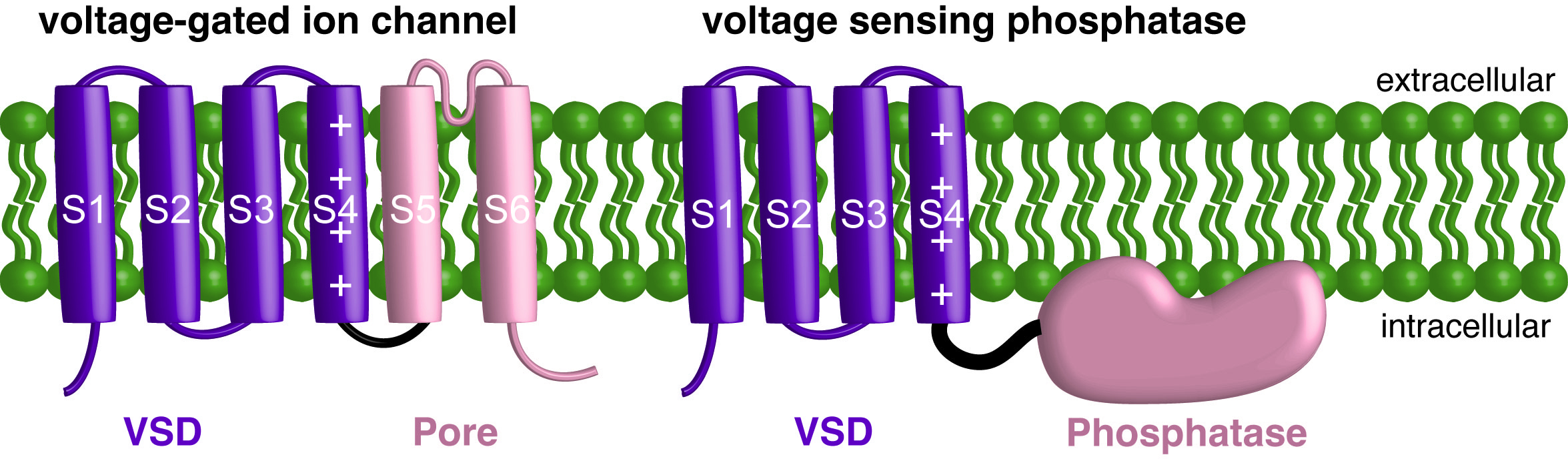
Figure 1. Cartoons of a single subunit of a voltage gated ion channel and a single subunit of VSP. The individual helices of the VSD are labeled and the charge carrying arginines on S4 are also labeled.
The linker was initially named because it connects the VSD and phosphatase domain. It shares a phosphatidylinositol phosphate (PIP) binding motif found in many other proteins. We showed that a single mutation in the linker can remove the ability of the VSD to activate the enzyme (Figure 2) (3). Thus, the “on” signal must go through the linker to activate the catalytic domain. We also showed that independently decreasing PIP concentrations changed how the VSD moved. When the mutation in the linker was added, the VSD no longer changed motions when PIPs were depleted. This result supports the idea that PIP binding to the linker may regulate VSP function. Since VSP also changes PIP concentrations, this regulation means that a feedback loop is generated when VSP activates.
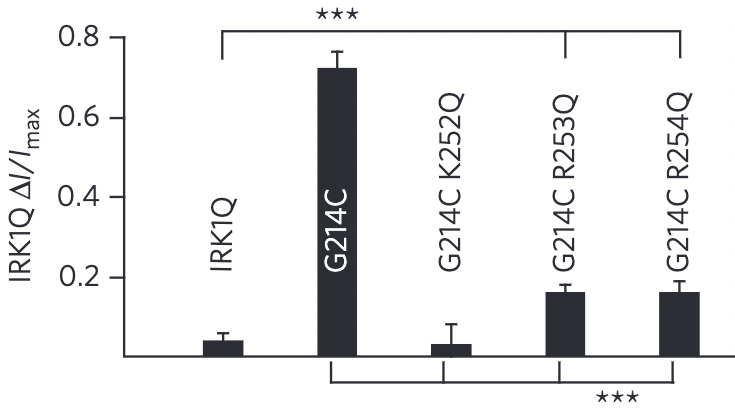
Figure 2. VSP activity as measured using the ion channel IRK. Linker mutations dramatically decreased (R253Q and R254Q) or completely eliminated (K252Q) any measurable activity as compared to wild type (G214C).
We worked with collaborators, Dr. Lijun Liu and Dr. Dan Minor (UCSF) to solve several crystal structures of the VSP phosphatase domain. They solved seven different structures, in several confirmations, with and without a soluble substrate (Figure 3) (4). With these various structures, we discovered that a particular loop in the catalytic domain appears to flip in and out of the active site to modulate catalytic activity (4). Mutating key residues in the loop dramatically increased activity or eliminated it all together depending on the amino acid being introduced (Figure 4). This loop is conserved in many phosphatases indicating that the vast amount of research on phosphatases also applies to VSP.
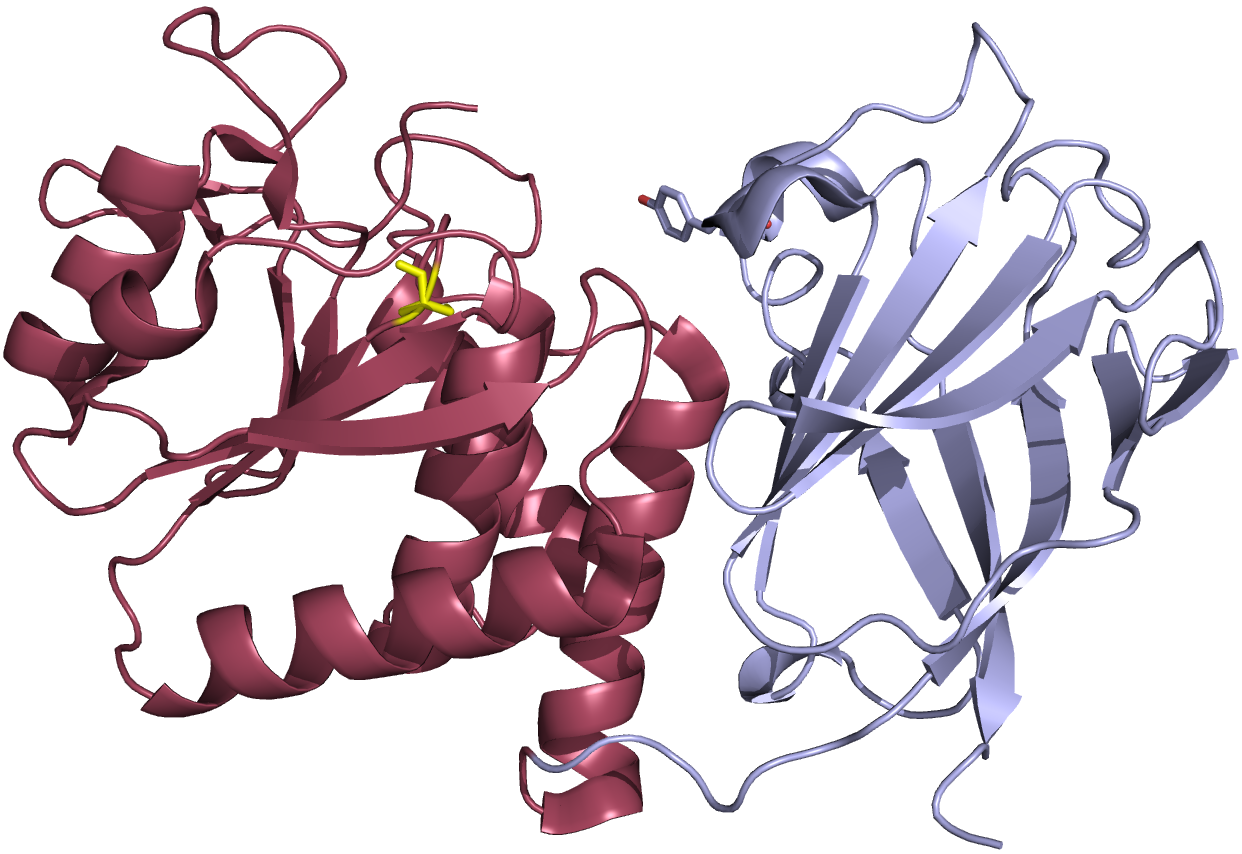
Figure 3. Crystal structure cartoon representation of the VSP phosphatase domain. The catalytic domain is show in red and the C2 domain is shown in gray. The active site cysteine (mutated to a serine for the purposes of the crystallization) is shown in yellow.
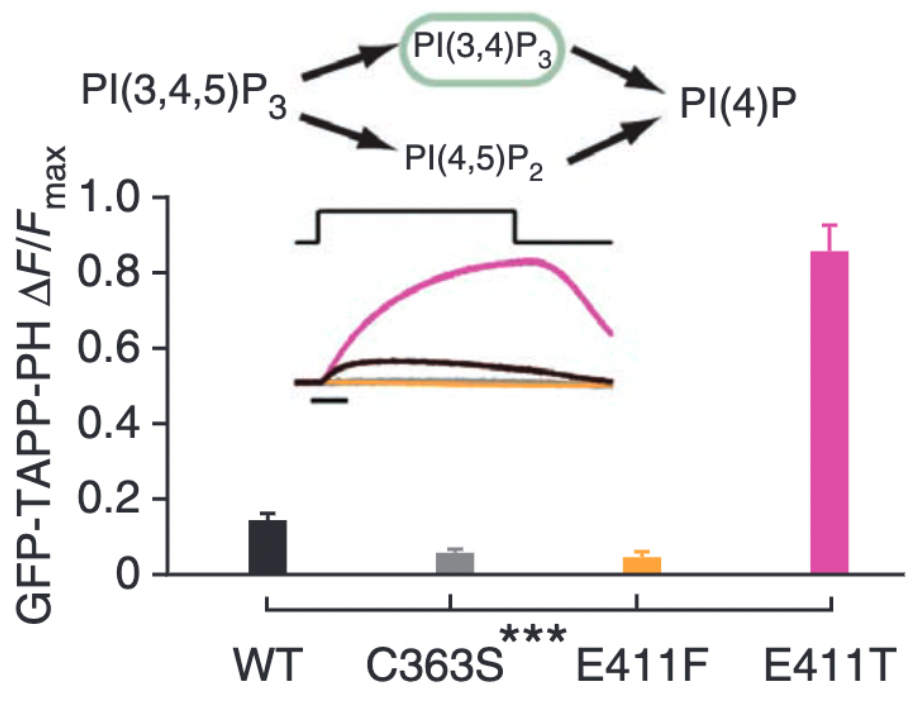
Figure 4. VSP activity as measured by the fluorescent biosensor, GFP-TAPP-PH. This biosensor monitors PI(3,4)P2 levels in particular so an increase in signal indicates more PI(3,4)P2 while a decrease in signal indicates less PI(3,4)P2. The bar graphs show how a E411F substitution eliminates activity compared to wild type (WT) while the E411T substitution dramatically increases activity compared to WT.
Interestingly, VSP has a C2 domain along side the catalytic domain. C2 domains are classic membrane binding domains and in general are not involved in catalysis. However, the structures we found showed a potential interaction between a C2 domain amino acid and the active site of the catalytic domain (Figure 5) (4). Further investigations into that region found that the C2 domain clearly influences catalytic activity as well as VSD activation (5). Removing the C2 domain entirely completely eliminated the 3-phosphatase activity and severely diminished the 5-phosphatase activity (Figure 6). These are very unexpected functions for a C2 domain and further indicate how VSP continues to defy our expectations.
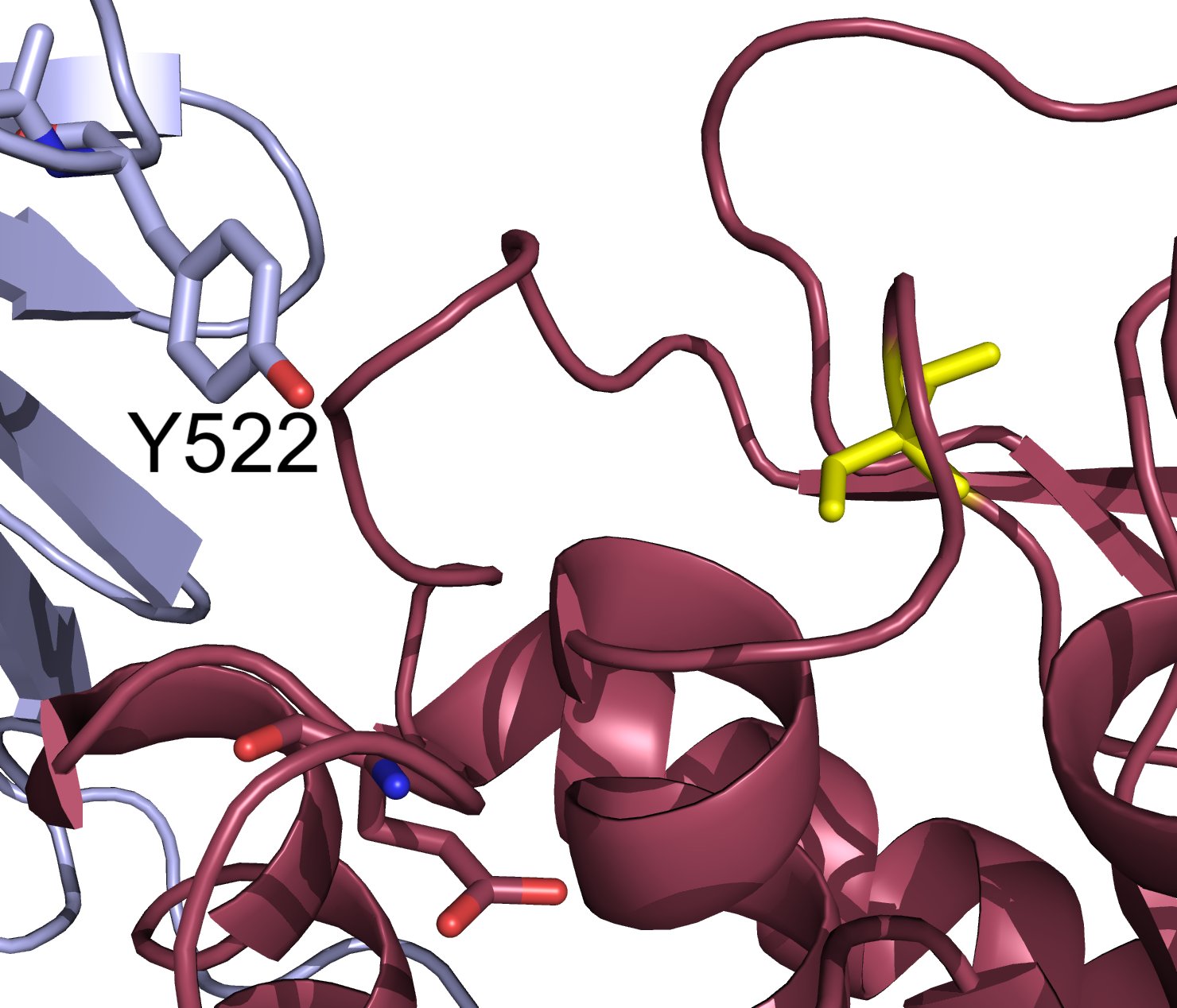
Figure 5. Zoom in on the crystal structure cartoon representation of the VSP phosphatase domain. The catalytic domain is show in red and the C2 domain is shown in gray. Labeled reside Y522 from the C2 domain is facing into the active site space. Catalytic site residue in yellow.
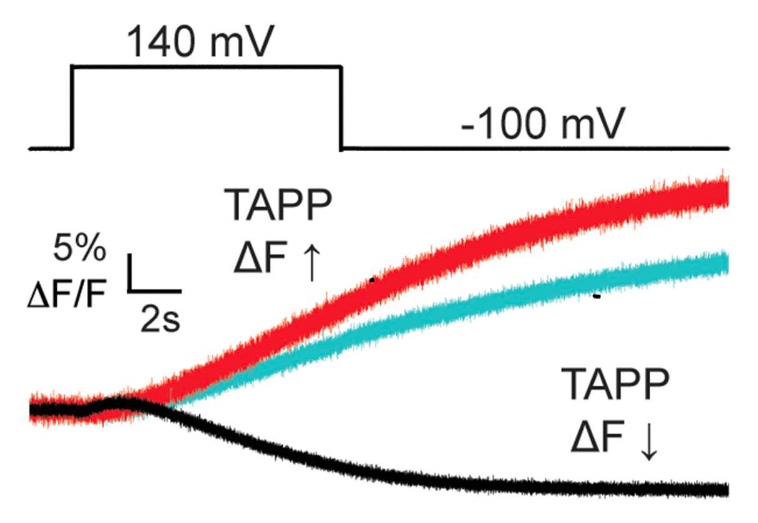
Figure 6. The GFP-TAPP-PH biosensor indicating PI(3,4)P2 levels in the cell. This biosensor monitors PI(3,4)P2 levels in particular so an increase in signal indicates more PI(3,4)P2 while a decrease in signal indicates less PI(3,4)P2. WT VSP (black) shows an initial increase and then decrease in fluorescence indicating robust 5- and 3-phosphatase activity. VSP with the C2 domain deleted (red) shows only an increase indicating some 5-phosphatase activity and no 3-phosphatase activity. Endogenous VSP (cyan) shows only an increase. VSP without the C2 domain is only a bit larger than the endogenous levels indicating reduced 5-phosphatase activity.
One of our first findings on VSP was that it could function as a single protein (monomer) as opposed to channels that typically need 2 or more units for their biological function (1). Subsequent research appeared to contradict our original finding, so we set out to re-explore whether VSP worked as a single protein or if multiple proteins work together. We discovered that indeed, VSP can work in pairs (dimers) (6). This conversion between monomers and dimers appears to be concentration dependent (Figure 7). Subsequent modeling showed that the changes in the voltage dependent activity did not need to consider dimerization to recapitulate the voltage dependent changes observed at different concentrations (7). However, mixing inactive with active proteins changed which reactions were catalyzed suggesting dimerization does impact the function of VSP (Figure 8) (6).
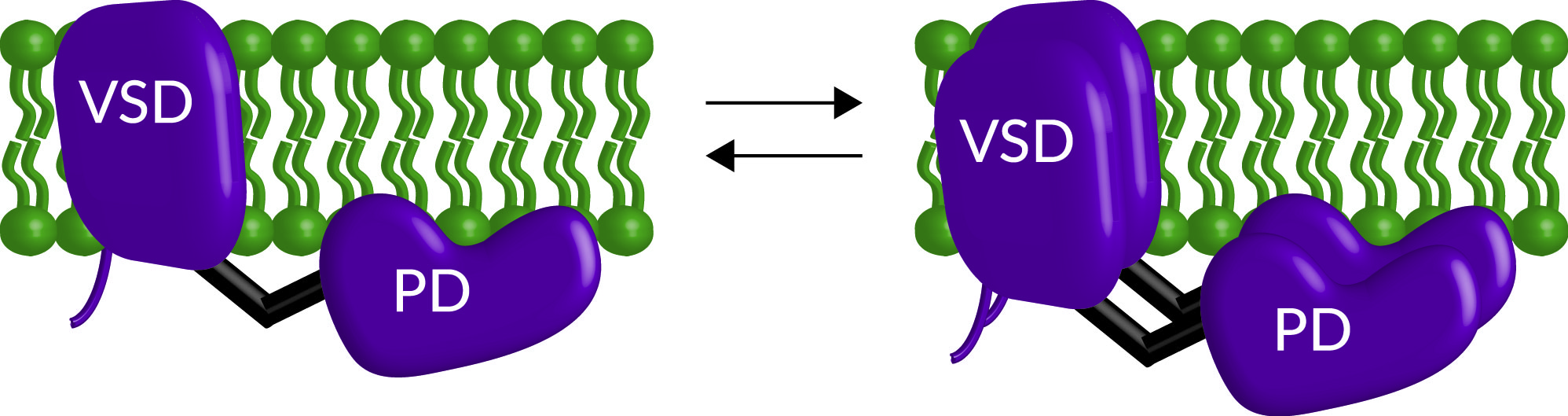
Figure 7. Cartoon representation of VSP dimerization. Both monomers and dimers exist on the membrane with higher concentrations of protein favoring more dimer formation.
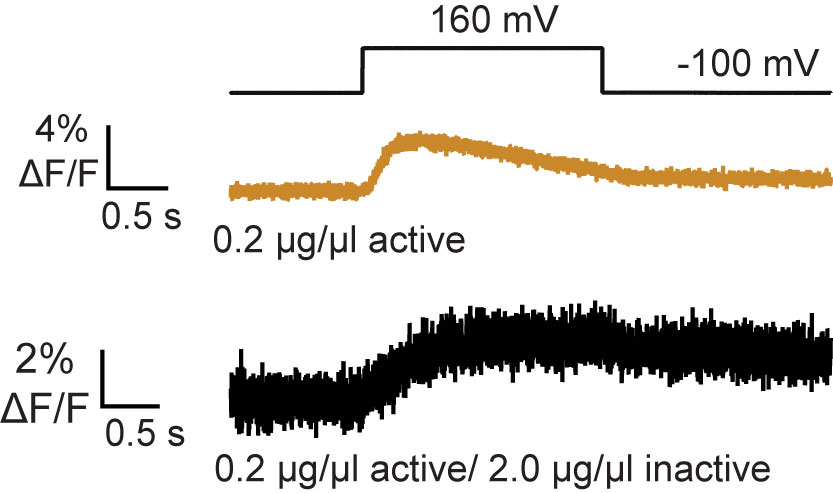
Figure 8. VSP activity as measured by the Förstor resonance energy transfer (FRET)-TAPP biosensor. Active/active dimers (tan) show an increase and decrease in signal indicating robust 5- and 3-phosphatase activity respectively. Active/inactive dimers (black) show only an increase in signal indicating no 3-phosphatase activity. These results show that dimer formation changes VSP activity.
Our most recent publication shows our investigation into how the VSD influences enzymatic activity, particularly focusing on S1 (8). This first helix of the domain is often overlooked and our data suggests that it plays a fairly large role in regulating VSP since even just a single mutation, L137A, dramatically changes both the kinetics and voltage dependence of activity. Dr. Kohout was interviewed and the research was highlighted by the journal for their Research News section (9).
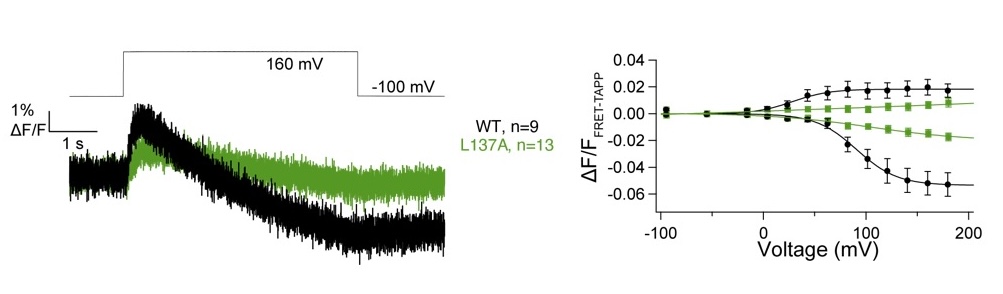
Figure 9. FRET-TAPP data for the L137A mutation compared to wild type. Both show an increase and decrease in FRET indicating both 5- and 3-phosphatase activity, however L137A is slower to activate both reactions (left) and requires significantly more energy (higher voltage) to activate (right).
We continue to address many of the questions that remain unanswered regarding VSP! What is the mechanism behind dimerization changing VSP catalytic specificity? How does PIP modulation impact VSP activation and inactivation? Under what circumstances does the cell require voltage regulated PIP concentrations? And more!
References
1. Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol 2008;15:106–8. https://doi.org/10.1038/nsmb1320. PubMed PMID: 18084307.
2. Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, et al. Closing in on the resting state of the Shaker K(+) channel. Neuron 2007;56:124–40. https://doi.org/10.1016/j.neuron.2007.09.023. PubMed PMID: 17920020.
3. Kohout SC, Bell SC, Liu L, Xu Q, Minor DL Jr, Isacoff EY. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat Chem Biol 2010;6:369–75. https://doi.org/10.1038/nchembio.349. PubMed PMID: 20364128; PubMed Central PMCID: PMC2857593.
4. Liu L, Kohout SC, Xu Q, Müller S, Kimberlin CR, Isacoff EY, et al. A glutamate switch controls voltage-sensitive phosphatase function. Nat Struct Mol Biol 2012;19:633–41. https://doi.org/10.1038/nsmb.2289. PubMed PMID: 22562138; PubMed Central PMCID: PMC3529583.
5. Castle PM, Zolman KD, Kohout SC. Voltage-sensing phosphatase modulation by a C2 domain. Front Pharmacol 2015;6:63. https://doi.org/10.3389/fphar.2015.00063. PubMed PMID: 25904865; PubMed Central PMCID: PMC4389355.
6. Rayaprolu V, Royal P, Stengel K, Sandoz G, Kohout SC. Dimerization of the voltage-sensing phosphatase controls its voltage-sensing and catalytic activity. J Gen Physiol 2018;150:683–96. https://doi.org/10.1085/jgp.201812064. PubMed PMID: 29695412; PubMed Central PMCID: PMC5940254.
7. Kruse M, Kohout SC, Hille B. Reinterpretation of the substrate specificity of the voltage-sensing phosphatase during dimerization. J Gen Physiol 2019;151:258–63. https://doi.org/10.1085/jgp.201812260. PubMed PMID: 30622132; PubMed Central PMCID: PMC6363406.
8. Rayaprolu, Vamseedhar, Heini M. Miettinen, William D. Baker, Victoria C. Young, Matthew Fisher, Gwendolyn Mueller, William O. Rankin, et al. 2024. “Hydrophobic Residues in S1 Modulate Enzymatic Function and Voltage Sensing in Voltage-Sensing Phosphatase.” The Journal of General Physiology 156 (7). PMID: 38771271 PMCID: PMC11109755
9. Short, Ben. 2024. “The S1 Helix Is a VIP in VSP.” The Journal of General Physiology 156 (7). https://doi.org/10.1085/jgp.202413612.