5'-UTRs
5'-UTRs
5'-UTRs
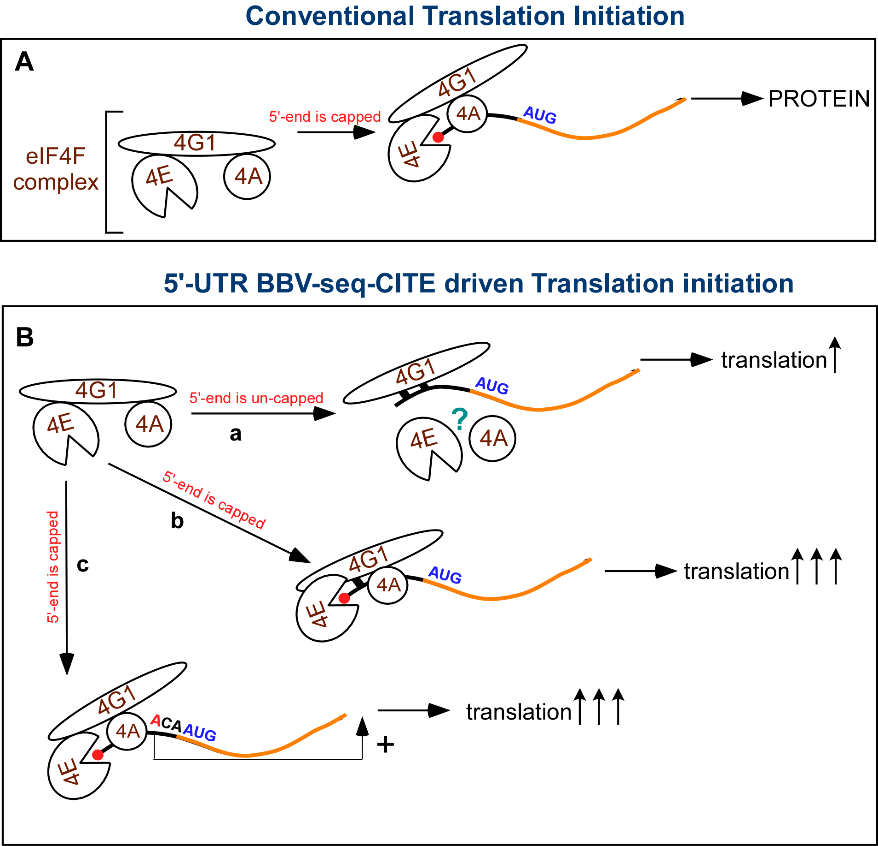
Translation initiation is one of the most critical regulatory steps in protein synthesis, and untranslated regions at the 5ʹ end of RNA (5ʹ-UTRs) play a key role in this process. 5ʹ-UTRs are non-coding regions within mRNAs that are located directly upstream of open reading frames (ORF). Undoubtedly, the most important function of 5ʹ-UTRs is the recruitment of multiple translation initiation factors required for ribosomal subunit association at the correctly selected AUG start codon. 5ʹ-UTRs not only provide the platform for cap-dependent translation initiation, but also possess multiple regulatory functions as modulators of translation initiation, ultimately contributing to the expression of a particular protein at a particular time in a particular environment. Our lab studies the mechanism by which a short, insect virus-derived sequence (BBV-seq, 39 nt) promotes cap-independent 5ʹ-end-dependent translation initiation.
We developed a yeast-based, cell-free translation system (Trainor et al., 2021-a) that can be “charged” with an mRNA of choice, while the production of translated proteins can be detected and quantified using various techniques. The yeast-based system is a powerful tool, as it allows the generation of translationally competent yeast extracts from genetically modified yeast strains, facilitating our understanding of the role of a particular gene. Using these cell-free translation reactions, we found that BBV-seq 1) does not constitute an IRES; 2) works as a Kozak element in the context of capped mRNAs, increasing the efficiency of translation initiation; 3) and functions as a previously undescribed cap-independent translation enhancer (CITE) on uncapped mRNAs. We propose that the BBV-seq-CITE element recruits translation initiation factor eIF4G1 to an uncapped mRNA to efficiently position the 40S subunit for scanning and identifying the start codon of the downstream gene (Trainor et al., 2021-b).
Future studies. This project opens a new arena for further investigations. To unravel details of the BBV-seq-mediated, cap-independent translation initiation, we are focused on the following questions:
- What domains of eIF4G1 are involved in direct BBV-seq binding?
- How does binding of eIF4G1 to BBV-seq 5ʹ-UTR affect eIF4A helicase recruitment?
- How does the BBV-seq CITE mechanism function on poly-adenylated RNAs?