Quality Control
Quality Control
Quality Control
What are the mechanisms of ribosome degradation and ribosome surveillance?
The process of ribosome biogenesis is well studied, but the mechanisms underlying ribosome degradation are still poorly defined. As of today, two major pathways of ribosome decay have been described: non-functional ribosome decay (NRD), which eliminates defective ribosomes; and ribophagy, a form of autophagy that removes ribosomes in bulk upon starvation.
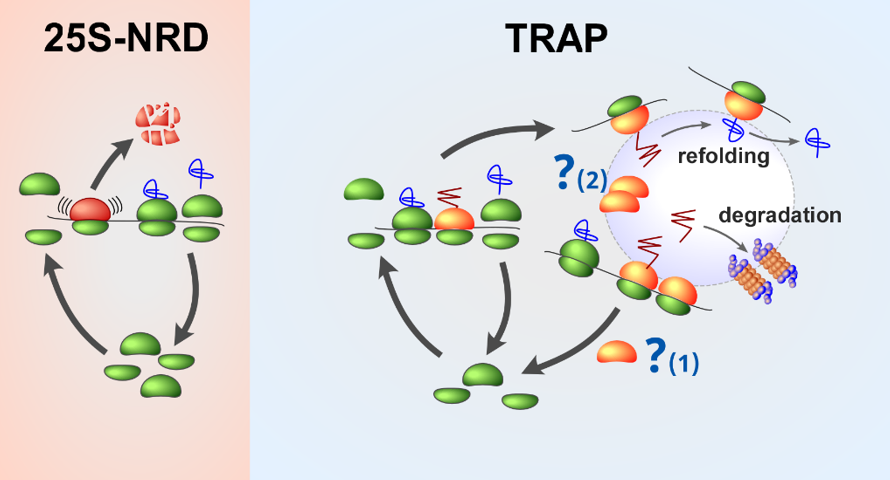
In our lab, we explore additional models of ribosome degradation that allow cells to recognize and remove old ribosomes and maintain a healthy ribosome pool, important for error-free translation and successful adaptation to a constantly changing environment. We have found that the E3 ubiquitin ligase Rsp5 is required for rRNA integrity by ubiquitinating ribosomal proteins on functional ribosomes (Shcherbik 2011). We have also demonstrated that suppressing TOR signaling induces a distinct cytoplasmic form of ribosome degradation in yeast (Pestov 2012). Both Rsp5-dependent and rapamycin-induced cytoplasmic turnover of ribosomes involves nucleases and is mechanistically distinct from ribophagy and NRD (Pestov 2012). Finally, we discovered a novel ribosome surveillance pathway, translational re-localization with aberrant polypeptides (TRAP), which operates on ribosomes during proteotoxic stress by sequestering ribosome-nascent chain complexes into perinuclear compartments (JUNQ) shared with misfolded proteins (Ghosh et al., 2020). We propose that TRAP represents a cellular strategy to maintain healthy protein homeostasis.
Future studies. To know more about TRAP, we are interested in addressing the following questions:
- What additional molecular factors operate within the TRAP pathway?
- Are TRAPed ribosomes released from JUNQ to participate in a new round of translation, or do they undergo degradation? In the latter case, what are the mechanism(s) that mediate degradation of faulty TRAPed ribosomes?
- What molecular aberrations (within mRNA or nascent polypeptide chains) activate TRAP?
How does co-translational protein quality control (QC) operate in cells?
Co-translational protein QC is a critical part of protein homeostasis, as up to 15% of all newly generated polypeptides in eukaryotic cells fail to fold or assemble properly. Recent studies have shown that cells begin to eliminate faulty polypeptides as soon as during translation. These co-translational QC systems are extremely important for proper protein homeostasis. Dysfunctional co-translational QC may cause the accumulation of aberrant polypeptides that contribute to neurodegeneration and cancer.
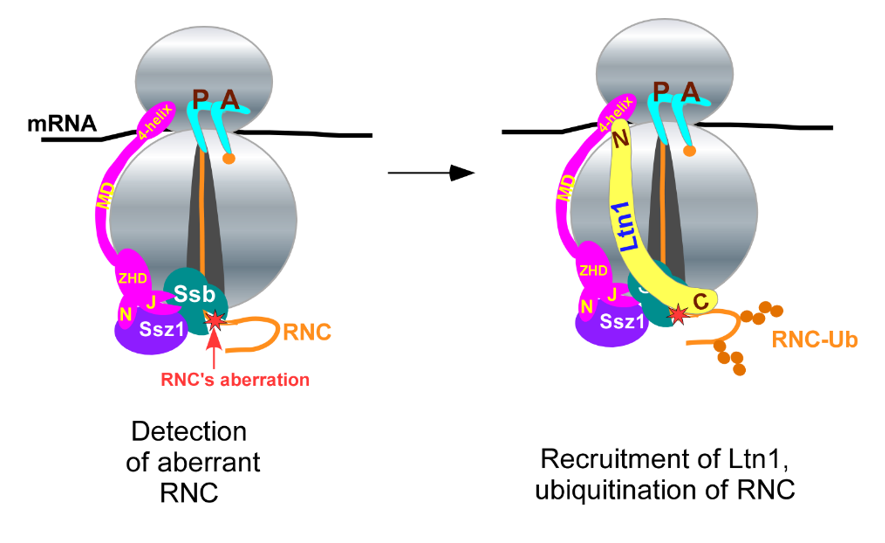
The most studied co-translational QC mechanism relies on enzymatic complexes of the ubiquitin-proteasome system (UPS) and operates on peptidyl-tRNAs associated with 60S ribosomal subunits. Additional co-translational QC mechanisms appear to exist, but they have received limited experimental attention. In our lab, we developed a biochemical approach to better track endogenous substrates of co-translational QC (Shcherbik, 2016). We demonstrated that QC of nascent chains can also take place on fully assembled ribosome 80S. In this pathway, which we termed 80S-RQC, the ribosome-selective chaperone triad Ssb/RAC facilitates positioning of Ub-ligase Ltn1 for efficient polypeptide modification (Ghosh and Shcherbik, 2020). We propose that this unconventional pathway constitutes a branch of co-translational QC for efficient and timely elimination of aberrant nascent polypeptide chains.
Future studies. There are unanswered questions that we are eager to address. They include:
- Deciphering molecular details of Ssb/RAC and Ltn1 interaction with ribosomes and determining whether Ssb/RAC-mediated accommodation of Ltn1 on a ribosome depends on different conformations of Hsp70-Ssb, i.e., ATP- or ADP-bound Ssb;
- Identifying substrates of 80S-RQC to determine the types of aberrations that trigger Ssb/RAC-dependent and Ssb/RAC-independent nascent polypeptide chain QC