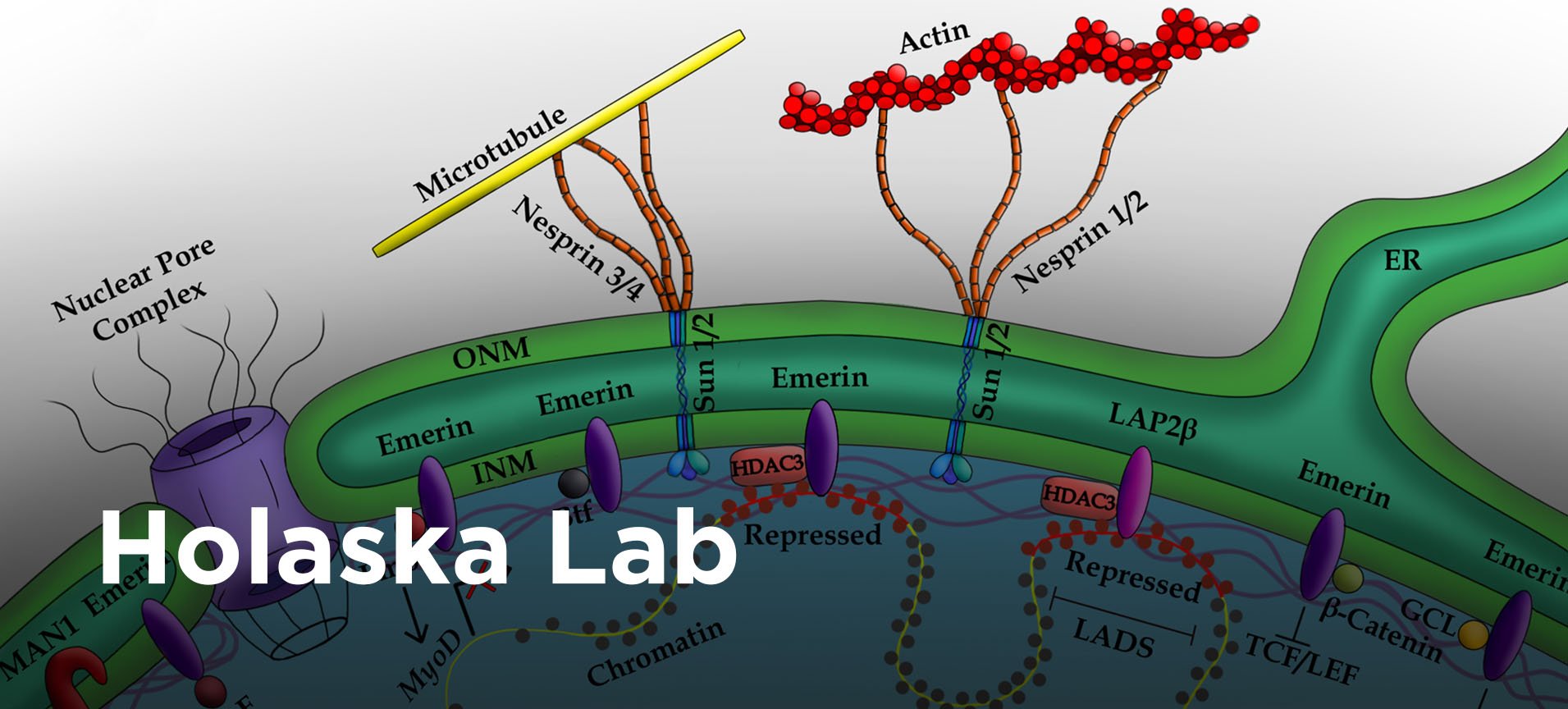
Publications
Publications
Publications (h-index: 24; i10-index: 27)
Complete List of Published Work in MyBibliography: https://www.ncbi.nlm.nih.gov/sites/myncbi/james.holaska.1/bibliography/41490064/public/?sort=date&direction=ascending.
Peer-reviewed publications in the primary literature, exclusive of abstracts:
- Holaska JM and Paschal BM. (1998) A cytosolic activity distinct from Crm1 mediates nuclear export of protein kinase inhibitor in permeabilized cells. Proc Natl Acad Sci USA 95, 14739-14744. http://www.pnas.org/content/95/25/14739.long
- Black BE, Levesque L, Holaska JM, Wood TC, and Paschal BM. (1999) Identification of an NTF2-related factor that binds RanGTP and regulates nuclear protein export. Mol Cell Biol 19, 8616-8624. http://mcb.asm.org/content/19/12/8616.long
- Lindsay ME, Holaska JM, Welch K, Paschal BM, and Macara IG. (2001) Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 153, 1391-1402. http://jcb.rupress.org/content/153/7/1391.long
- Black BE, Holaska JM, Rastinejad F, and Paschal BM. (2001) DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr Biol 11, 1749-1758. http://www.cell.com/current-biology/retrieve/pii/S0960982201005371
- Black BE, Holaska JM, Levesque L, Ossareh-Nazari B, Dargemont C, and Paschal BM. (2001) NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J Cell Biol 152, 141-155. http://jcb.rupress.org/content/152/1/141.long
- Holaska JM, Black BE, Love DC, Hanover JA, Leszyk J, and Paschal BM. (2001) Calreticulin is a receptor for nuclear export. J Cell Biol 152, 127-140. http://jcb.rupress.org/content/152/1/127.long
- Brownawell AM, Holaska JM, Macara IG, and Paschal BM. (2002) The use of permeabilized cell systems to study nuclear transport. Meth Mol Biol 189, 209-229. http://www.springerlink.com/content/q41133j682500850/#section=82121&page=1
- Holaska JM, Black BE, Rastinejad F, and Paschal BM. (2002) Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol 22, 6286-97. http://mcb.asm.org/content/22/17/6286.long
- Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. (2002) Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett 525, 135-40. http://www.sciencedirect.com/science/article/pii/S0014579302031058
- Wilkinson FL, Holaska JM, Zhang Z, Sharma A, Manilal S, Holt I, Stamm S, Wilson KL, and Morris GE. (2003) Emerin interacts in vitro with the splicing-associated factor, YT521-B. Eur J Cell Biol 270, 2459-66. http://onlinelibrary.wiley.com/doi/10.1046/j.1432-1033.2003.03617.x/full
- Holaska JM, Lee KK, Kowalski AK, and Wilson KL. (2003) Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem 278, 6969-75. http://www.jbc.org/content/278/9/6969.long
- Haraguchi T, Holaska JM, Yamane M, Koujin T, Hashiguchi N, Mori C, Wilson KL, and Hiraoka Y. (2004) Emerin binding to Btf, a death-promoting transcriptional repressor, is disrupted by a missense mutation that causes Emery-Dreifuss muscular dystrophy. Eur J Cell Biol 271, 1035-1045. http://onlinelibrary.wiley.com/doi/10.1111/j.1432-1033.2004.04007.x/full
- Holaska JM, Kowalski AK, and Wilson KL. (2004) Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear envelope. PloS Biol 2, 1354-1362. http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0020231
- Holaska, JM and Wilson KL. (2006) Lmo7, an emerin-binding protein that shuttles between the cell surface and nucleus and regulates emerin transcription. Hum Mol Genet 15, 3459-3472. http://hmg.oxfordjournals.org/content/15/23/3459.long
- Holaska, JM and Wilson KL. (2007) An emerin proteome: purification of distinct emerin-containing complexes from HeLa cells suggests a molecular basis for diverse roles including gene regulation, mRNA splicing, signaling and nuclear architecture. Biochemistry 46, 8897-8908. http://pubs.acs.org/doi/abs/10.1021/bi602636m
- Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, Pytel P, McNally E. (2008) Disruption of nesprin-1 produces an Emery Dreifuss Muscular Dystrophy-like phenotype in mice. Hum Mol Genet 18, 607-620. http://hmg.oxfordjournals.org/content/18/4/607.long
- Puckelwartz MJ, Kessler EJ, Kim G, DeWitt MM, Zhang Y, Earley JU, Depreux FF, Holaska JM, Mewborn SK, Pytel P, and McNally EM (2010). Nesprin-1 mutations in human and murine cardiomyopathy. J Mol Cell Cardiol 48, 600-608. http://www.sciencedirect.com/science/article/pii/S0022282809004817
- Dedeic Z, Cetera M, Cohen TV, Holaska JM (2011). Emerin inhibits Lmo7 binding to the Pax3 and MyoD promoters and expression of myoblast differentiation genes. J Cell Sci 124, 1691-1702. http://jcs.biologists.org/content/124/10/1691.long
- Demmerle J, Koch AJ, Holaska JM (2012). The nuclear envelope protein emerin binds to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J Biol Chem 287, 22080-22088. http://www.jbc.org/content/287/26/22080.long
- Koch AJ, Holaska JM (2012). Loss of emerin alters myogenic signaling and miRNA expression in mouse myogenic progenitors. PLoS One 7(5):e37262. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0037262
- Demmerle J, Koch AJ, Holaska JM (2013). The interaction between emerin and HDAC3 is required for proper genomic reorganization during muscle stem cell differentiation. Chrom Res 21, 765-79. http://link.springer.com/article/10.1007%2Fs10577-013-9381-9
- Mull A, Kim G, Holaska JM (2015). LMO7-null mice exhibit phenotypes consistent with Emery-Dreifuss muscular dystrophy. Muscle Nerve 51, 222-8. http://onlinelibrary.wiley.com/doi/10.1002/mus.24286/full
- Depreux FF, Puckelwartz MJ, Augustynowicz A, Wolfgeher D, Labno CM, Pierre-Louis D, Cicka D, Kron SJ, Holaska JM, McNally EM (2015). Disruption of the lamin A and matrin-3 interaction by myopathic LMNA mutations. Hum Mol Genet 24, 4284-95. http://hmg.oxfordjournals.org/content/24/15/4284.long
- Collins CM, Ellis J, Holaska JM (2017). MAPK signaling pathways and HDAC3 activity are disrupted during emerin-null myogenic progenitor differentiation. Dis Model Mech 10, 385-97. doi: 10.1242/dmm.028787. http://dmm.biologists.org/content/early/2017/02/09/dmm.028787.long
- Iyer A, Koch AJ, Holaska JM (2017). Expression profiling of differentiating emerin-null myogenic progenitor identifies molecular pathways implicated in their impaired differentiation. Cells, 6 (4), 38. doi:10.3390/cells6040038. http://www.mdpi.com/2073-4409/6/4/38/htm
Peer-reviewed book chapters and review articles:
- Holaska JM, Wilson KL, and Mansharamani M. (2002) The nuclear envelope, lamins, and nuclear assembly. Curr Opin Cell Biol 14, 357-64. http://www.sciencedirect.com/science/article/pii/S0955067402003290
- Mansharamani M, Wilson KL, and Holaska JM. (2003) Dynamics of the Vertebrate Nuclear Envelope. In: Dynamics of Nuclear Envelope Assembly in Embryos and Somatic Cells. P. Collas, ed. (Landes Bioscience). http://www.springerlink.com/content/nu2l36126g218232/
- Holaska, JM and Wilson KL. (2006) Multiple roles for emerin: implications for Emery-Dreifuss muscular dystrophy. Anat Rec A Discov Mol Cell Evol Biol 288, 676-80. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2559942/?tool=pubmed
- Holaska JM. (2008) Emerin and signaling via the nuclear lamina. Circ Res 103, 16-23. http://circres.ahajournals.org/content/103/1/16.long
- Koch AJ and Holaska JM (2014). Emerin in health and disease. Semin Cell Dev Biol 29, 95-106. Invited review. http://www.sciencedirect.com/science/article/pii/S1084952113001377
- Holaska JM (2016). Diseases of the nucleoskeleton. Comp Phys, 15, 1655-1674. Invited review. http://onlinelibrary.wiley.com/doi/10.1002/cphy.c150039/pdf
- Collins CM, Nee KA, Holaska JM (2016). The nuclear envelope protein emerin and its interacting proteins. eLS,1-9. Invited review.
- Holaska JM (2017). Emerin and the making of muscle. Research Features 104, 42-45. Invited article focusing on my research. http://researchfeatures.com/2017/02/01/emerin-and-the-making-of-muscle/
Works in review, in preparation, etc. Not yet publically available
- Bossone K, Ellis J and Holaska JM (2019). Inhibiting histone acetyltransferase activity rescues differentiation of emerin-null myogenic progenitors. Submitted
- Koch AJ, Collins CM, Nee KA and Holaska JM (2019). Emerin-dependent dynamic changes in genomic organization during myogenic differentiation cause large-scale changes in the transcriptome: implications for muscle disease. Manuscript in preparation
- Liddane AG, Campbell M, Mercier I and Holaska JM (2019). Emerin inhibits invasive breast cancer cell migration, invasion and metastasis by increasing nuclear size and structure through its interactions with the nucleoskeleton and chromatin. Manuscript in preparation.