Part II
Part II
Emerin Functions in Genomic Organization to Regulate the Coordinated Temporal Expression of Genes Driving Stem Cell Differentiation
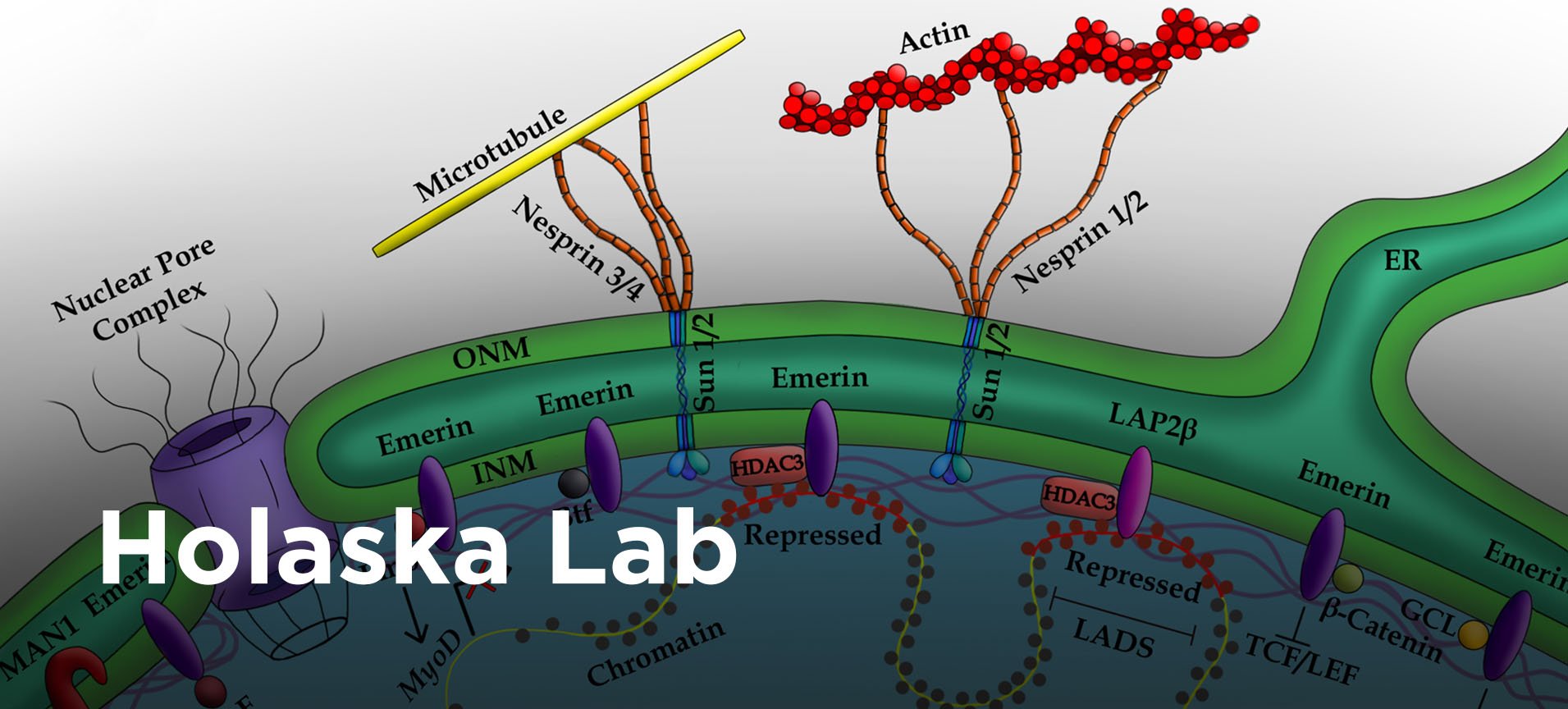
![]() The field lacks a fundamental understanding of the mechanisms underlying how proteins at the nuclear envelope regulate transcription factor activity and genomic organization. Further, our understanding of how genomic organization at the nuclear periphery is initiated and maintained to establish repressed domains at the nuclear lamina and active domains at nuclear pore complexes is particularly lacking. Understanding emerin function in nuclear architecture, gene expression and cell signaling during skeletal muscle stem cell differentiation will not only answer fundamental questions regarding how the nuclear envelope regulates genomic architecture and gene expression, but it will also contribute significantly to elucidating the EDMD mechanism. Understanding emerin regulation of genomic architecture will also impact other diseases resulting from altered chromatin architecture.
The field lacks a fundamental understanding of the mechanisms underlying how proteins at the nuclear envelope regulate transcription factor activity and genomic organization. Further, our understanding of how genomic organization at the nuclear periphery is initiated and maintained to establish repressed domains at the nuclear lamina and active domains at nuclear pore complexes is particularly lacking. Understanding emerin function in nuclear architecture, gene expression and cell signaling during skeletal muscle stem cell differentiation will not only answer fundamental questions regarding how the nuclear envelope regulates genomic architecture and gene expression, but it will also contribute significantly to elucidating the EDMD mechanism. Understanding emerin regulation of genomic architecture will also impact other diseases resulting from altered chromatin architecture.
Current studies are designed to identify specific molecular interactions that determine genomic organization at the nuclear lamina and how this organization is altered during muscle stem cell differentiation. These studies will be among the first to investigate the mechanism for recruitment, propagation or maintenance of repressive chromatin at the nuclear envelope by the nuclear lamina. To date, studies focused mainly on defining the nuclear lamina as a repressive environment, but failed to analyze the mechanism underlying repression by the nuclear lamina. How the genome is dynamically reorganized at the nuclear envelope to regulate the coordinated expression of differentiation genes will also be examined. Molecular pathways involved in the impaired myogenic differentiation associated with EDMD will be uncovered by these studies. These studies will further our understanding of the mechanisms regulating stem cell differentiation and how this fails in muscle disease.
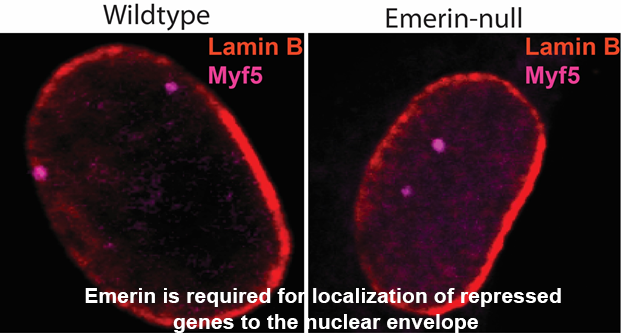 One of the emerin-binding complexes predicted to mediate genomic localization to the nuclear periphery is the NCoR complex, a chromatin repressive complex containing histone deacetylate 3 (HDAC3). Since the initial identification of this emerin-interacting complex, we made substantial progress in understanding the interaction between emerin and the HDAC3 complex. We found emerin binds directly to HDAC3 and stimulates its deacetylase activity. The interaction between emerin and HDAC3 is required for localizing repressed myogenic differentiation genes to the nuclear periphery. These genomic regions associate with emerin when repressed during myogenesis. Upon activation, they are released from emerin and move to the nuclear interior. Importantly, nuclear envelope localization of repressed genomic loci was dependent on emerin and HDAC3. This localization also required the interaction of HDAC3 with emerin. Additionally, loss of emerin in myogenic progenitors caused significant changes in the expression of key myogenic differentiation genes, demonstrating that alterations in genomic architecture affect gene expression. These studies established a new model for emerin regulation of repressive chromatin formation at the nuclear periphery: emerin binding to and activation of HDAC3 at the nuclear envelope stabilizes the initial events of chromatin repression (deacetylation), which serves to recruit other repressive complexes to the nuclear envelope to further repress chromatin and establish a stable, transcriptionally repressive environment.
One of the emerin-binding complexes predicted to mediate genomic localization to the nuclear periphery is the NCoR complex, a chromatin repressive complex containing histone deacetylate 3 (HDAC3). Since the initial identification of this emerin-interacting complex, we made substantial progress in understanding the interaction between emerin and the HDAC3 complex. We found emerin binds directly to HDAC3 and stimulates its deacetylase activity. The interaction between emerin and HDAC3 is required for localizing repressed myogenic differentiation genes to the nuclear periphery. These genomic regions associate with emerin when repressed during myogenesis. Upon activation, they are released from emerin and move to the nuclear interior. Importantly, nuclear envelope localization of repressed genomic loci was dependent on emerin and HDAC3. This localization also required the interaction of HDAC3 with emerin. Additionally, loss of emerin in myogenic progenitors caused significant changes in the expression of key myogenic differentiation genes, demonstrating that alterations in genomic architecture affect gene expression. These studies established a new model for emerin regulation of repressive chromatin formation at the nuclear periphery: emerin binding to and activation of HDAC3 at the nuclear envelope stabilizes the initial events of chromatin repression (deacetylation), which serves to recruit other repressive complexes to the nuclear envelope to further repress chromatin and establish a stable, transcriptionally repressive environment.
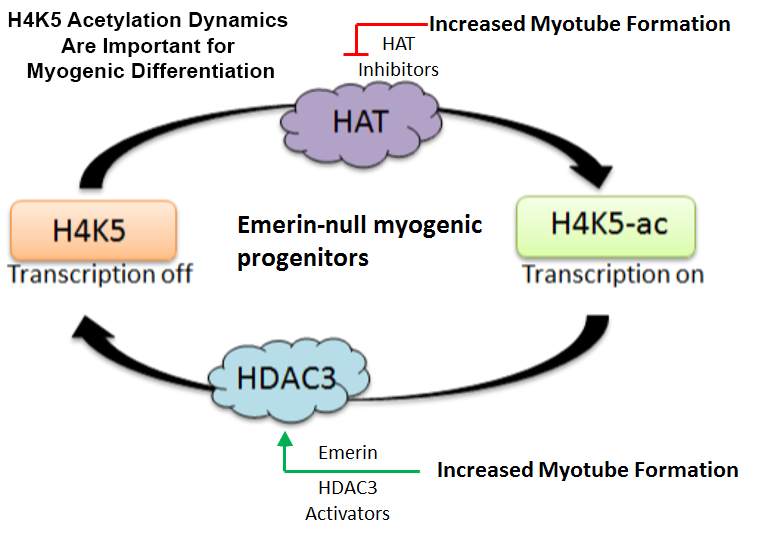 HDAC3 preferentially deacetylates lysine 5 of histone 4 (H4K5), suggesting H4K5 acetylation plays an important role in myogenic differentiation. Using histone acetyltransferase (HAT) inhibitors specifically targeting HATs that preferentially modify H4K5, we found H4K5 acetylation levels must be precisely regulated to allow differentiation to proceed normally. This was the first demonstration of H4K5 acetylation dynamics being important for myogenic regulation. We propose emerin acts as a rheostat to precisely control H4K5ac levels. Future studies will test this model.
HDAC3 preferentially deacetylates lysine 5 of histone 4 (H4K5), suggesting H4K5 acetylation plays an important role in myogenic differentiation. Using histone acetyltransferase (HAT) inhibitors specifically targeting HATs that preferentially modify H4K5, we found H4K5 acetylation levels must be precisely regulated to allow differentiation to proceed normally. This was the first demonstration of H4K5 acetylation dynamics being important for myogenic regulation. We propose emerin acts as a rheostat to precisely control H4K5ac levels. Future studies will test this model.