Part III
Part III
Identification and Characterization of Molecular Pathways Disrupted in Muscle Disease: Drug Discovery and Development
 We showed emerin-null myogenic progenitors fail to transcriptionally reprogram upon differentiation induction, which we predict is caused by the inability of emerin-null cells to reorganize the genome. Supporting this hypothesis, our previous studies showed histone deacetylase 3 (HDAC3) was an important regulator of myogenic differentiation through regulation of H4K5ac levels. The goal of this research project is to identify molecular pathways important for myogenic differentiation and implicated in the EDMD disease mechanism. Multiple approaches are being used for these studies. The first approach is to screen small molecules for rescuing emerin-null differentiation using medium-throughput, microscopy-based screens. The second approach is to use RNA sequencing to identify gene networks disrupted amongst five different emerin mutants that cause disease. We predict molecular pathways implicated in EDMD will be those containing genes altered in all emerin mutants.
We showed emerin-null myogenic progenitors fail to transcriptionally reprogram upon differentiation induction, which we predict is caused by the inability of emerin-null cells to reorganize the genome. Supporting this hypothesis, our previous studies showed histone deacetylase 3 (HDAC3) was an important regulator of myogenic differentiation through regulation of H4K5ac levels. The goal of this research project is to identify molecular pathways important for myogenic differentiation and implicated in the EDMD disease mechanism. Multiple approaches are being used for these studies. The first approach is to screen small molecules for rescuing emerin-null differentiation using medium-throughput, microscopy-based screens. The second approach is to use RNA sequencing to identify gene networks disrupted amongst five different emerin mutants that cause disease. We predict molecular pathways implicated in EDMD will be those containing genes altered in all emerin mutants.
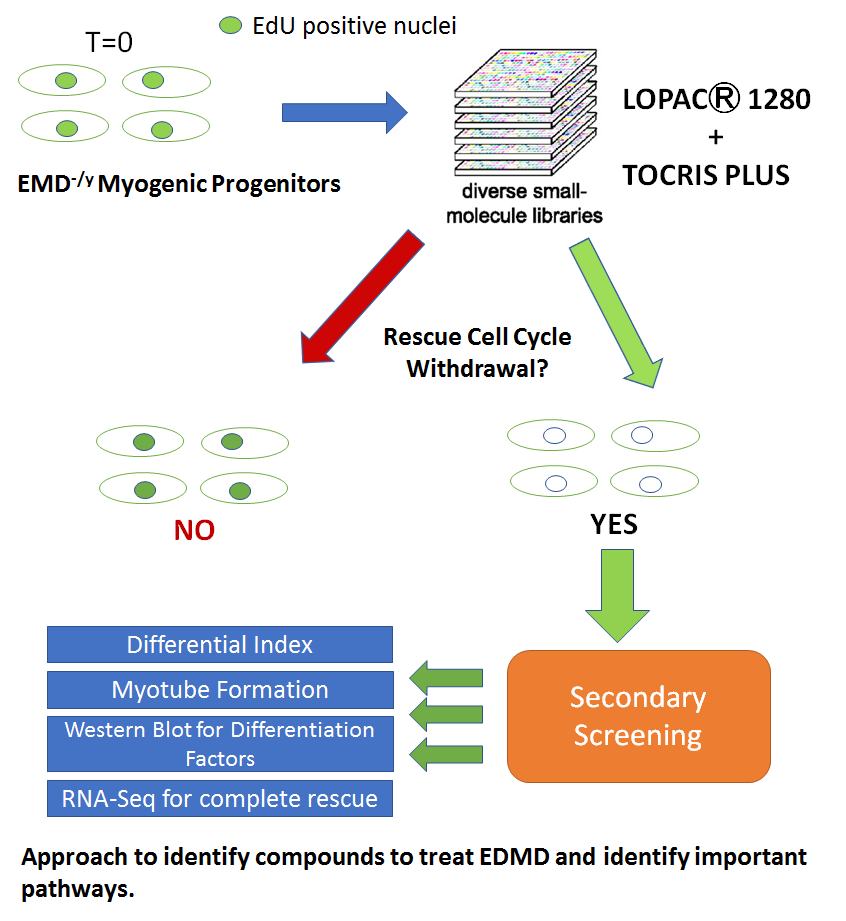
 The first approach will screen small molecules for rescuing emerin-null myogenic differentiation with the goal of identifying putative drugs and the molecular targets for treating EDMD. The proposed studies are designed to identify new small molecules that rescue emerin-null myogenic differentiation. Molecular pathways involved in the impaired myogenic differentiation associated with EDMD will be uncovered by the proposed studies. These studies will identify candidate compounds for treating the skeletal muscle defects of EDMD. Future studies would test these molecules in EDMD mouse models and patient myoblasts. These studies will have a broad impact on muscle disease, as they will further our understanding of the mechanisms regulating stem cell differentiation, including the role of nuclear envelope proteins. These studies also have implications for acute and chronic muscle injury, which rely on muscle regeneration after injury.
The first approach will screen small molecules for rescuing emerin-null myogenic differentiation with the goal of identifying putative drugs and the molecular targets for treating EDMD. The proposed studies are designed to identify new small molecules that rescue emerin-null myogenic differentiation. Molecular pathways involved in the impaired myogenic differentiation associated with EDMD will be uncovered by the proposed studies. These studies will identify candidate compounds for treating the skeletal muscle defects of EDMD. Future studies would test these molecules in EDMD mouse models and patient myoblasts. These studies will have a broad impact on muscle disease, as they will further our understanding of the mechanisms regulating stem cell differentiation, including the role of nuclear envelope proteins. These studies also have implications for acute and chronic muscle injury, which rely on muscle regeneration after injury.
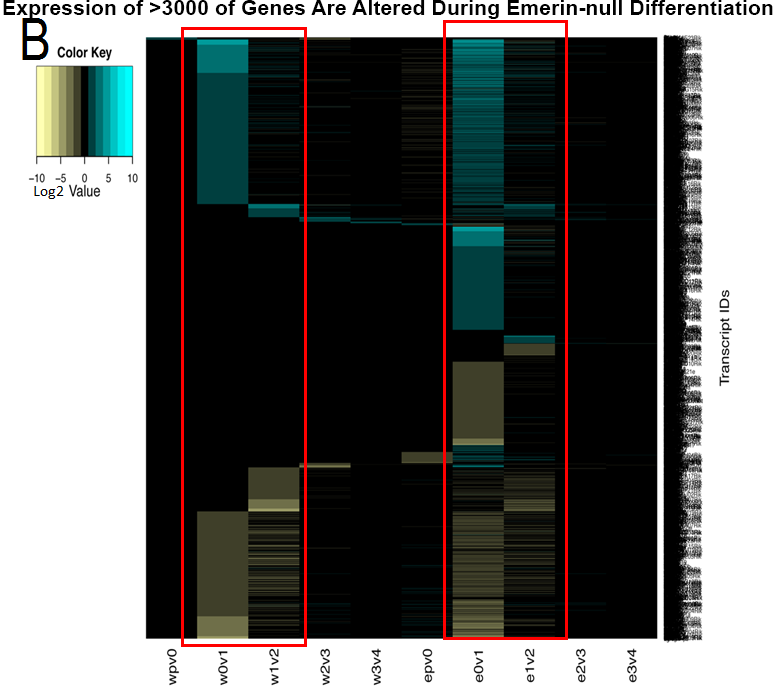
 In the second approach, we monitored mRNA, miRNA and protein expression in emerin-null myogenic progenitors during differentiation to begin deciphering the molecular mechanisms underlying how loss of emerin causes impaired myogenic differentiation. Approximately 3,500 genes showed altered expression during differentiation of emerin-null myogenic progenitors. Bioinformatics mapped these gene products to molecular pathways. Many of these genes mapped to Wnt, IGF-1, TGF-b and Notch signaling pathways, as expected. These signaling pathways are important for myogenic differentiation and disruption of these pathways causes impaired myogenic differentiation in mice and humans. Importantly, a large number of new pathways were identified. These new pathways function in the cell cycle, in muscle cell differentiation and development, in muscle cell function, and in MAPK signaling.
In the second approach, we monitored mRNA, miRNA and protein expression in emerin-null myogenic progenitors during differentiation to begin deciphering the molecular mechanisms underlying how loss of emerin causes impaired myogenic differentiation. Approximately 3,500 genes showed altered expression during differentiation of emerin-null myogenic progenitors. Bioinformatics mapped these gene products to molecular pathways. Many of these genes mapped to Wnt, IGF-1, TGF-b and Notch signaling pathways, as expected. These signaling pathways are important for myogenic differentiation and disruption of these pathways causes impaired myogenic differentiation in mice and humans. Importantly, a large number of new pathways were identified. These new pathways function in the cell cycle, in muscle cell differentiation and development, in muscle cell function, and in MAPK signaling.
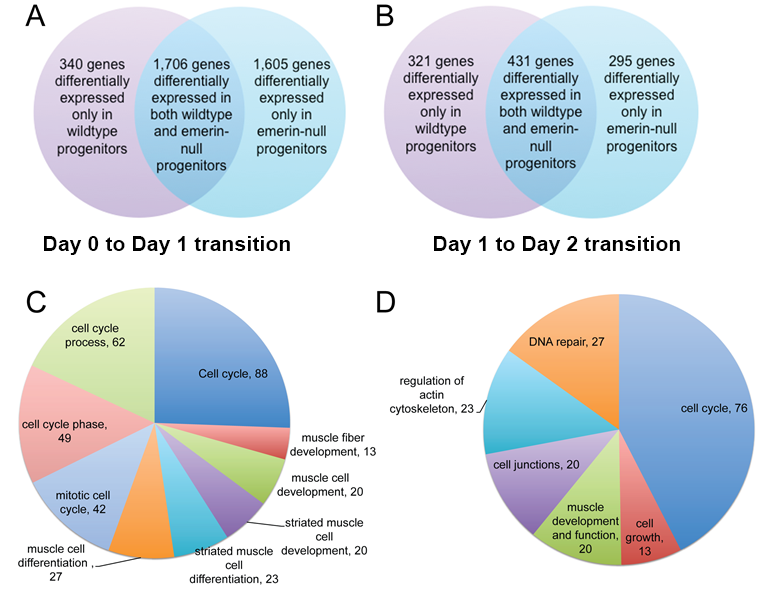
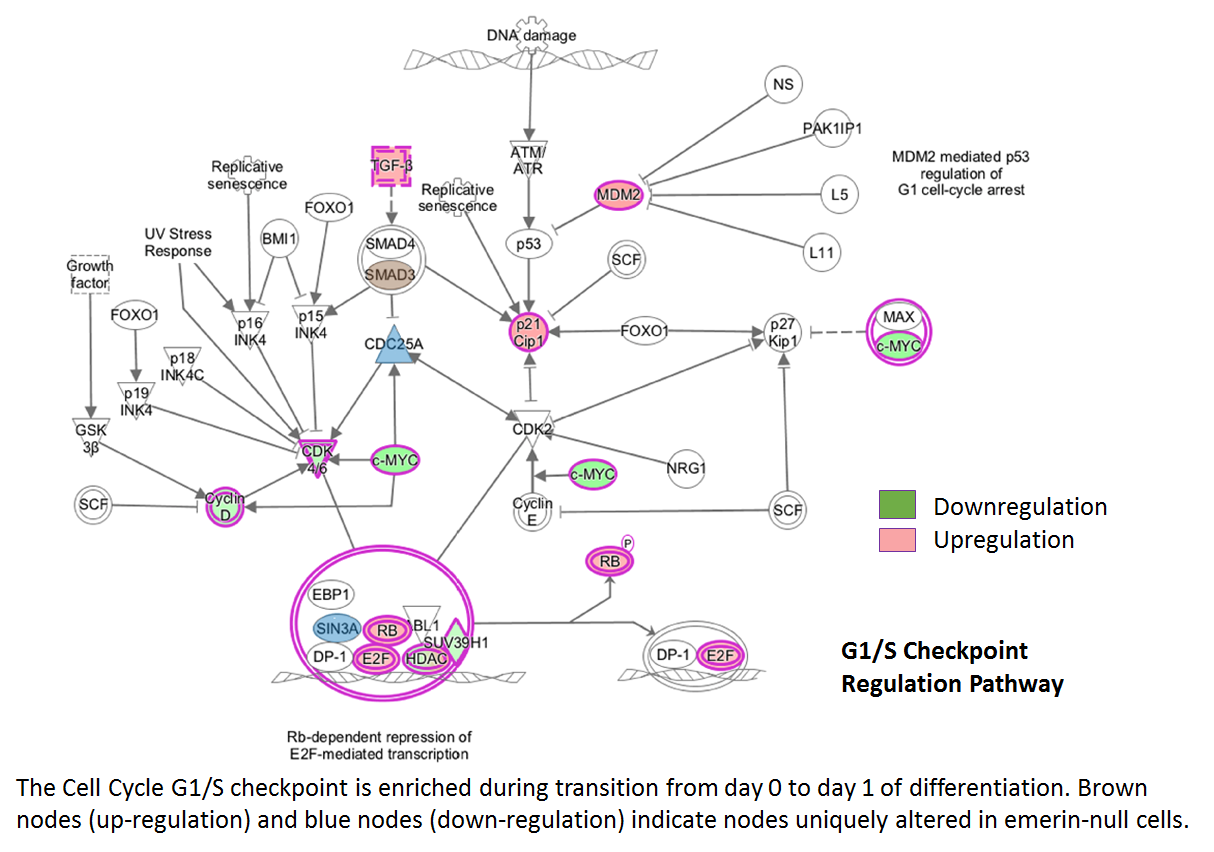 Our preliminary analysis validated many of these genes and confirmed signaling through these pathways was disrupted in differentiating emerin-null myogenic progenitors. Small molecule inhibitors and activators of selected pathways showed different pathways preferentially act at specific transition points during differentiation, as some inhibitors specifically rescued cell cycle withdrawal, while others rescued myoblast fusion or myotube maturation. We anticipate identifying small molecules that activate or block molecular pathways important for the EDMD disease mechanism. These small molecules would then be tested in animal models of EDMD in future experiments.
Our preliminary analysis validated many of these genes and confirmed signaling through these pathways was disrupted in differentiating emerin-null myogenic progenitors. Small molecule inhibitors and activators of selected pathways showed different pathways preferentially act at specific transition points during differentiation, as some inhibitors specifically rescued cell cycle withdrawal, while others rescued myoblast fusion or myotube maturation. We anticipate identifying small molecules that activate or block molecular pathways important for the EDMD disease mechanism. These small molecules would then be tested in animal models of EDMD in future experiments.
| ⇐ Back to Part II | Part IV ⇒ |